Using photoaffinity probes based on a motif found in kinase-targeting anticancer drugs, researchers from KU Leuven have shown that their off-targets are not only found in the kinase families, but also in other proteins. They have published their findings in Communications Chemistry.
A common way to study enzymes is to use activity-based probes. These small molecules contain a reactive moiety that mimics the substrate of an enzyme. Depending on the probe, you can form a covalent bond between it and the enzyme. If you attach a molecular label to the probe, you can then filter out the enzyme for analysis.
‘But sometimes there are enzymatic mechanisms for which mechanism-based inhibitors do not apply’, says Steven Verhelst, professor of chemical biology at KU Leuven. In this case, photoaffinity-based probes could be used. ‘These get around the problem by binding non-covalently to the enzyme. A few years ago, our group started to look at various targets, including kinases.’
It was around this time that Verhelst recruited Dimitris Korovesis as a postdoctoral fellow, who already had experience in the synthesis and application of kinase inhibitors and imaging probes.
Kinase dysfunction can lead to several types of cancer, for which kinase inhibitors can act as drugs. ‘But as with all drugs, you want to know how specific they are and whether there are other proteins they target’, says Korovesis, who is now a postdoctoral researcher at the Institute of Molecular Biology and Biotechnology in Crete.
Trivial
Korovesis started out with an experimental molecule – KIRA6 – that was designed to target a specific protein, IRE1α. Another group knew the molecule had an off-target, but they didn’t know which one. ‘We found what they were looking for, but what was interesting about KIRA6 is that it has a very common structural core that matches several other kinase inhibitors.’
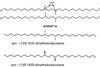
That’s when Korovesis and Verhelst began to wonder: what small differences between these very similar molecules might make them more selective? With this in mind, they selected a few known kinase inhibitors and synthesised some of their own, all containing the same imidazopyrazine core structure, to convert them all into photoaffinity labels and look at their selectivity.
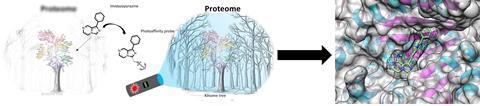
During the synthesis, which was partly carried out by master’s student Christel Mérillat, the team encountered a number of hurdles and setbacks. ‘At several points we had to switch between protecting groups, for instance because we saw that the one we had chosen fell off during purification’, says Korovesis. Another example is the protection of an aromatic amine in the pyrazine ring. ‘According to the literature, you shouldn’t have to protect it to do a coupling reaction, but in our hands it wouldn’t work and we had to protect it to make the diazirine.’ Verhelst adds: ‘Choosing your protecting groups can sometimes seem trivial, but this situation shows that this isn’t always the case.’
Incomplete
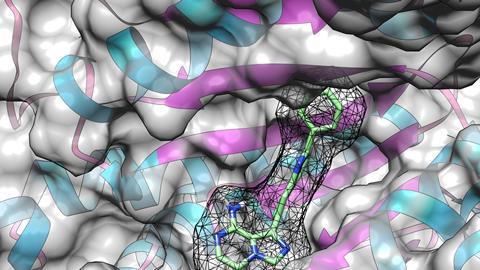
It’s important to take a broader view of off-target profiles in drug discovery and development, says Verhelst. ‘You shouldn’t just look at the selectivity of kinase inhibitors within the kinome [the family of kinase enzymes], but within the wider proteome. There are different techniques to assess the selectivity of kinase inhibitors, but some of them only look at the kinome itself, which makes the determination of selectivity incomplete without also looking at the other ATP and nucleotide binding proteins that we have found as off-targets.’
If you find these off-targets early on, you won’t get unexpected surprises later on in the drug discovery process. ‘And I’m not suggesting that you have to use photoaffinity labelling, as long as you’re looking at the whole proteome. I think that’s the main message of this manuscript’, concludes Verhelst.
Korovesis, D. et al. (2025) Commun Chem 8(34), DOI: 10.1038/s42004-025-01436-y










Nog geen opmerkingen