Despite the fact that lignin has been analysed for quite some time, it is still a considerable challenge to map the branches properly. An American-Flemish team found a mass spectrometry method that has made this a whole lot easier.
Lignin, a main component of the cell walls of plant tissues such as wood, is an aromatic polymer that is partly linear and partly branched. It is composed mainly of guaiacyl and syringyl units, but can also contain other types of phenolic units. If you want to know the shape of such a polymer, you can break it into pieces and analyse it with mass spectrometry (MS). ‘Analysing lignin with MS has been going on for quite a long time’, says Kris Morreel, visiting professor at UGent and senior scientist at RIC Group. ‘But some oligomeric fragments that occur infrequently are difficult to identify. Yet that analysis is important to pinpoint branches in the sequence.’ Naturally, if you don’t know what you’re dealing with, you can’t do anything with it.
The identity of most lignin chunks can be identified with LC-MS, but dimers originating from branching points are trickier. ‘That is because those dimers have difficult to break bonds’, Morreel explains. ‘These are 5-5 and 4-O-5, a biphenyl with a covalent C-C bond and a biphenyl ether, respectively. Both bonds prefer not to break.’
Morreel worked with a team of US scientists to elucidate the fragments of precisely those dimers with a special form of MS. ‘We compared data from quadrupole ion trap [QIT, ed.] with quadrupole time-of-flight [QTOF, ed.] mass spectrometry which work slightly differently.’ (See box QTOF vs QIT below.) In short, the QTOF variant gave more fragments and thus more data. ‘Ultimately, this allowed us to distinguish between 5-5 and 4-O-5, where previously this was not possible. Indeed, we saw that 4-O-5 does fragment, but 5-5 does not. We can thus incorporate that knowledge in the future to find out a branching pattern.’
With this method, you can generate more and more data on lignin, including branching patterns. Morreel: ‘You can now use this work to use algorithms to start looking at branching points of longer oligolignols, i.e. longer lignin chains. If we then build a database of identified oligolignols and then run machine learning on them, you can work towards some guidelines for interpreting the data.’
Dong, X., Mayes, H., Morreel, K. et al. (2022) ChemSusChem e202201441, doi.org/10.1002/cssc.202201441
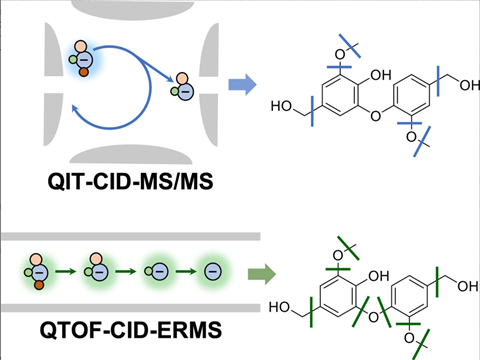
QTOF vs. QIT
QTOF can be compared to a pair of interconnected chambers where the different steps take place. The parent ion that enters the collision cell from the first cell must have enough energy to leave it again, so that daughter ions can be analysed in the third cell. This way, you still get a relatively large amount of fragmentation.
Whereas you can call QTOF a tandem in space, QIT is a tandem in time. It is a kind of egg-shaped cavity into which ions enter. Once that egg has filled, the ions are pushed out. There is less fragmentation and therefore less data, but the advantage is that you can always isolate one ion and fragment it further, giving you info on daughter and granddaughter ions.
You don’t get that information easily from a QTOF, you have to derive it yourself from the data you collect. Indeed, that further fragmentation does happen, but all at once. So QIT and QTOF are actually complementary. Want a lot of information? Then use QTOF. If you want to elucidate a structure by focusing on the fragmentation mechanism, then QIT is sometimes better because you can really look at each ion individually.













Nog geen opmerkingen