In Bioconjugate Chemistry, Brussels-based researchers describe synthetic, nanobody-derived peptides that bind a top-priority pathogen. This could enable new ways to fight antibiotic-resistant bacteria.
Antibiotic resistance is a serious, fast-growing problem. New antimicrobials entering the market are sparse, while the number of multidrug-, extensively drug- and even pandrug-resistant bacterial strains is only increasing.
New strategies to treat these unruly pathogens are urgently needed, for example through focusing on small peptides (cell-penetrating peptides, CPPs, or antimicrobial peptides, AMPs) that can aspecifically bind to bacterial membranes, penetrate them or disrupt them via electrostatic interactions. It is precisely the aspecific nature of these peptides that makes them much less likely to induce resistance.
Extreme survivor
One of the most worrisome resistant pathogens, according to the World Health Organisation, is Acinetobacter baumannii. This Gram-negative bacterium is almost impossible to fight: it is resistant to a wide range of antibiotics, insensitive to disinfectants and can survive for a long time in extreme conditions. A. baumannii easily attaches to ventilators and catheters, posing a serious threat to the already vulnerable patients who depend on these devices.
Nanobody
Charles Van der Henst’s lab (VIB / VU Brussels) has now identified a nanobody, a ‘small’ antibody, which binds to the outer membrane of A. baumannii. Nanobodies have three complementarity-determining regions (CDRs); loop-like structures that ensure antigen recognition and binding, with CDR3 playing a dominant role. Together with the organic chemistry group of Steven Ballet (VUB), they decided to synthesize linear peptides based on the CDR3 sequence of this particular nanobody to see if they also bind to A. baumannii.
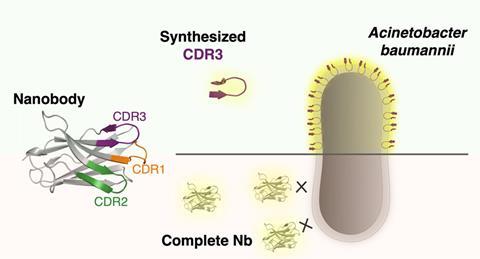
Bridge
This turned out to be a smart move; a linear analogue of CDR3 managed to bind to multiple, live A. baumannii isolates, unlike the complete nanobody that could only bind to fixed cells. To make the linear peptides more resemblant of loop-shaped CDRs and because that structure is likely to be important for robust recognition and binding, the team introduced a 1,2,3-triazole bridge between the terminal residues of the peptide. An additional advantage it that a cyclic structure makes the peptide less vulnerable to proteolytic degradation, resulting in longer-lasting activity. The cyclic versions also exhibited good binding affinity for A. baumannii and turned out to be more stable than the linear forms.
Even though these results cannot be immediately translated to a new antibiotic, but according to the authors, these peptides do offer an interesting option to serve as targets for, for example, peptide-drug conjugates to bind very specifically to bacterial cells. A key advantage of these small peptides over intact nanobodies is that they are easier and cheaper to produce.













Nog geen opmerkingen