In the battle against antibiotic resistance, you have to look for holdouts everywhere. A team from Umeå University in Sweden has been studying how bacteria pass on their resistance genes to each other. As well as gaining a better understanding of the regulation of the bacterial secretion system, it also resulted in a beautiful picture.
In this fantasy landscape, everything seems peaceful and bright, but the reality is grim. In this coloured image, taken by KNCV member Josy ter Beek using a scanning electron microscope, we see clumps of Enterococcus faecalis (the yellow spheres), a notorious ‘hospital bacterium’.
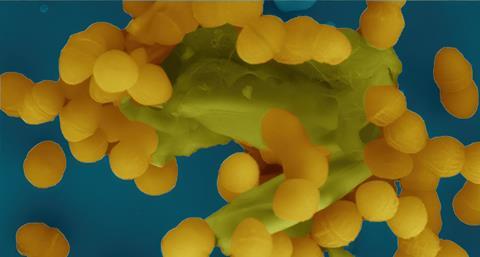
Resistance to antibiotics is one of the reasons why E. faecalis is so difficult to control. Ter Beek and her colleagues at Umeå University in Sweden are investigating how resistance spreads in E. faecalis. It was already known that type 4 secretion systems, T4SS, play a role in the transfer of (resistance) genes by acting as ‘copy machines’.
The enzyme PrgK is a crucial subunit of T4SS, as it cuts the bacterial cell wall so that the copied genetic material can be secreted. The Swedish study, published in mBio, now shows the role of the three components of PrgK in regulating this enzyme. By targeting one of these components, it might be possible to prevent the spread of resistance by T4SS.
Wei-Sheng Sun, et al., Breaking barriers: pCF10 type 4 secretion system relies on a self-regulating muramidase to modulate the cell wall, mBio (2024), doi:10.1138/mbio.0488-24











Nog geen opmerkingen