Chemists in Nijmegen have developed a reservoir computing system that uses the formose reaction to perform complex calculations, Nature reports. ‘You can use the same experimental data multiple times to perform hundreds of computational tasks.’
‘Since the early days of the computing era, people have thought about using molecules instead of transistors in computers because, at that time, they were smaller’, says Wilhelm Huck, professor of physical organic chemistry at Radboud University (RU). ‘But today there are disadvantages: transistors are now smaller than molecules, and a computer based on molecules works much slower than one based on electrons.’
That’s why the idea of molecular computing has moved on to DNA, where you can do calculations with smart circuits in the base network. Some time ago, for example, researchers showed that a whole series of DNA reactions could be used to calculate the square root of a given number.
‘We have simulated the carbon metabolism of E. coli with our formose reservoir’
‘But what we’re doing is entirely different, it doesn’t use normal computer architecture’, continues Huck. He refers to reservoir computing (RC), invented by German-born Herbert Jaeger, now a professor at the University of Groningen. RC deals with a weak point of computers, namely non-linear processes. ‘That is because you have to take very small steps over a long period of time to see what happens’, says Huck. ‘And that quickly becomes incredibly expensive. But it’s exactly those processes that you want use a computer for. Think of climate models or complex chemical reactions.’ RC offers a solution to this problem.
Automatic
But why does the molecular reservoir calculation work so well? ‘Because you don’t have to train the reaction we use, that is, the formose reaction’, Huck explains. ‘Every chemical reaction has its own characteristics, its own reaction constants. So the training is automatic.’
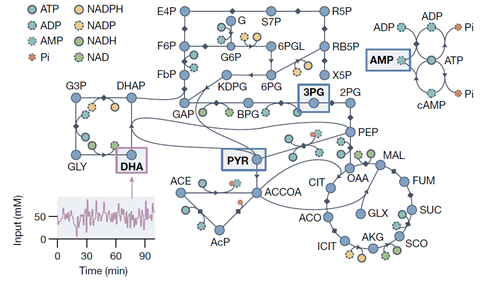
There are four types of input to the reservoir, which can be controlled via a flow system: formaldehyde, dihydroxyacetone, NaOH and CaCl2. The output is a mass spectrum, measured by an ion mobility MS. On the spectrum you can see 106 different ions, ’although not every ion is unique, as some traces are identical’, says Huck. So the output depends on the input, and you can then read out the non-linear response (the set of ions) by assigning different ’weights’ to the ion signals. You can use the same experimental data multiple times to perform hundreds of computational tasks by applying different sets of weights.
‘After a day of measurements, the whole MS instrument smelled like caramel’
Huck and colleagues provided a good example of what’s possible. Huck: ‘The reservoir can solve many mathematical equations at once. For example, we did an experiment where we simulated a model of the carbon metabolism of an E. coli bacterium with our formose reservoir.’ And it worked, even though the metabolism consists of 87 substrates and 92 reactions, which is quite a complex biochemical system.
Realisation
‘The fact that this is possible at all is quite extraordinary’, says Huck. ‘Normally you only see this in silico, and there are some examples in materials, but now that it has been shown to work with molecules, it opens the door to a very large group of chemical systems that can process large amounts of information.’
It has led to a realisation, says Huck. ‘There are many networks in a living cell. Until now, we have thought of these networks mainly as linear pathways, but perhaps chemical domains such as biological cells can be modelled much better by a reservoir computing system. The information that you put into the formose reservoir spreads across all dimensions of the network, which means that chemical systems that have a similarly meshed network structure will also have properties for processing information.’
Brown mess
The team first took samples using a GCMS, which measures a spectrum every few minutes. They measured thousands of spectra in this way, but this is far too slow for time series. ‘We then decided to inject our samples directly into Jana Roithová’s [Professor of Spectroscopy and Catalysis, RU, ed.] mass spectrometer without any pre-processing.’
That was quite a risk. ‘The formose reaction takes place with sugars and slowly forms caramel and finally black tar. So after a day of measurements, the whole instrument smelled of caramel and we had to remove this brown mess from the instrument. We thought the MS wouldn’t survive, but we’ve been doing it for a year and it doesn’t seem to cause any problems.’
‘Once it clicks, you see opportunities to apply reservoir computing everywhere’
To move this work towards application, Huck’s group is working with computer company IBM to build a reservoir computing reactor in a silicon chip. ‘With the same techniques used to make advanced computer chips, you should be able to make reactor chips’, explains Huck. ‘What we’re currently doing with pumps, tubes and MS needs to be scaled down to the microlitre scale so that you can put dozens of formose reactors on a chip. In the long run, this would allow you to solve serious computational problems.’
Pong
Their own research group is not standing still, either. ‘Internally, we are busy with another network that also works, but with formose we are moving towards a more autonomous system where you process the data from the MS in real time and use it to drive the pumps of your reactor. In other words, a self-driving system. PhD student Thijs de Jong has been working on this and is almost at the point where he can make the reservoir computer play Pong against itself, proving that the formose reaction is really a powerful computer. Once it clicks, you can see the possibilities of using reservoir computing everywhere.’
Baltussen, M.G. et al. (2024) Nature, DOI: 10.1038/s41586-024-07567-x












Nog geen opmerkingen