The ability of artificial intelligence to predict protein folding is a real game changer in structural biology, but the predictions have several shortcomings. NKI researchers use their own algorithm to overcome some of these shortcomings.
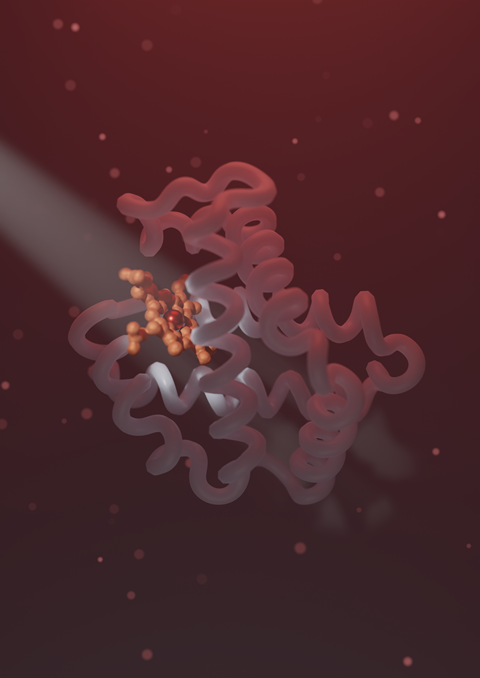
Determining protein structures required labour-intensive analysis in the laboratory until the advent of AlphaFold and RoseTTAfold in 2021. Both methods use artificial intelligence to predict protein structures based on amino acid sequence. The development has been hugely influential, but the models lack biological context and interpretation. AlphaFold predicts only one state, even though proteins are highly dynamic. In addition, the protein models lack ligands. For example, haemoglobin needs haem to adopt its structure, but haem is not present in the AlphaFold model. These shortcomings inspired researchers in the group led by Anastassis (‘Tassos’) Perrakis at the Netherlands Cancer Institute (NKI) to combine databases and add more information to the model.
Copy and paste
The first step was quick, says Ida de Vries, a PhD student in Perrakis’ group. Her colleague Maarten Hekkelman said he would see if he could fit the ligands into the protein model, and not long after he came back into her office: “I think it works! Hekkelman’s algorithm worked so quickly and efficiently that they decided to apply it to the entire AlphaFold database. The result is AlphaFill - an algorithm that uses amino acid sequence and structure similarity to certain experimentally determined structures to fit the missing ligands of those experimentally determined structures into the AlphaFold model. Alphafill compares sequences from AlphaFold models with protein sequences from structures in PDB-REDO, a supplement to the Protein Data Bank (PDB). If we find a hit, we superimpose the 3D structure of the AlphaFold model on the 3D structure corresponding to the sequence in PDB-REDO,’ explains De Vries. If there are important ligands such as small molecules or metal ions in the PDB-REDO model, we copy and paste them into the AlphaFold model.
Clash score
To validate the algorithm, De Vries only used proteins whose experimentally determined structure exactly matched the AlphaFold structure. The error rate is then equal to the error rate of the fold model. We cannot generate a validation for all ligands that we have inserted into an AlphaFold model, but we give two reliability scores in our model: one for the binding site and one for the clash score. The latter indicates whether the protein and ligand are too close together. AlphaFill shows the reliability for each ligand with a colour code: yellow for unreliable and red for very unreliable.
De Vries and colleagues published their validated database in Nature Methods in November 2022. The AlphaFill database now contains 1 million protein structures, all available at alphafill.eu. They created a web service where users can view the structures, request missing AlphaFold structures, and upload their own structures. By the beginning of February 2023, visitors had already uploaded 600 of their own structures and the web service had been visited by more than 8,000 people,’ says De Vries. We see researchers using the tool, even outside the NKI. She sees NKI researchers using AlphaFill to help them find binding sites. As an example, she cites a kinase in which AlphaFill glued both ATP and ADP into the model. This is possible because you cannot tell from the structural model which is which. The reliability scores allow us to say which of the two is most likely and whether it is the active or inactive variant of the kinase.
Addition
The current algorithm is limited by the structures in PDB-REDO. To overcome this limitation, the team is working on a way to predict the ligand-binding site of the protein. We started relatively simply with the metal ions,” explains De Vries. There is only one atom per ligand and they are important for catalysis and structural integrity. My PhD student Ren Xie has written the algorithm and it seems to work, but we are still extensively testing and validating it. The last two years have shown that AlphaFold is very useful but cannot replace structural biology experiments. Although AlphaFill is a nice addition, it also lacks the dynamics of real proteins in a realistic context. De Vries wants to understand how and why proteins undergo a conformational change when they bind to a partner protein. Even better would be to understand how a protein folds. But that is even more difficult. There are still many unanswered questions.












Nog geen opmerkingen